Omar Gadir, Ph. D., Founder/CEO, Iteru Systems
Stages Of Drug Discovery
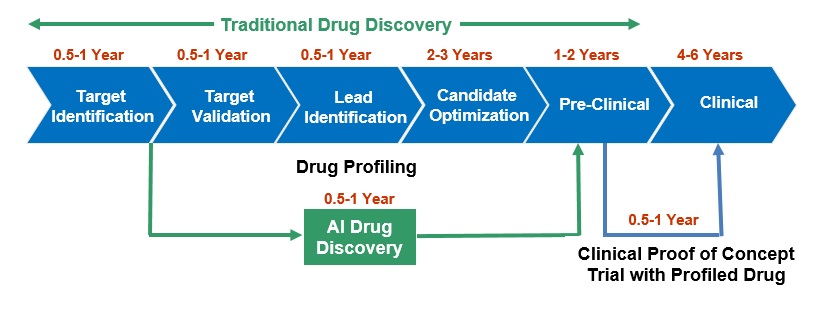
Fig. 1 AI shortens and reduces the cost of drug discovery
Traditional drug discovery is a daunting task. It takes on average 14 years and costs about $2.6B to develop a drug. Moreover, only about 5% of experimental drugs make it to market. The process is based on the costly and time-consuming trial and error. There is an urgent need for a change. One of the promising approaches is to adopt Artificial Intelligence (AI) in drug discovery. As shown in Fig. 1. AI can drastically cut down the expense of drug discovery and saves many years spent on drug profiling.
The most challenging aspect of drug discovery is cancer drug discovery. Thirty-five years after the ‘war on cancer’ was declared by President Nixon, the discovery of anticancer drugs remains a challenge. Recently, targeted small molecules and biologics established a new frontier for cancer drug discovery and treatment. AI can play a crucial role in reshaping this new frontier. For AI to succeed it should provide highly accurate results. This in turn depends on understanding biomedical data and the extracted features to be used in AI analysis.
Feature extraction and feature selection
Biomedical data is one of the most, if not the most complex data. Its complexity is covered in subsequent sections. It has its own specialized terminology and many acronyms. Moreover, there are complex interactions between hundreds of biological entities. While designing a feature extractor, a data scientist or a software engineer should look at the data from a pharma domain expert’s perspective. The person must have some understanding of what the domain expert looks for and how he/she interprets the data. This requires some knowledge of the internals of the data.
Iteru Systems recognized that prior to formulating a solution to the problem of feature extraction, we have to educate ourselves about biomedical data. Feature extraction is a critical step in obtaining accurate AI results. It should be designed to deal with the distinctive and unique features of biomedical data. This must be done at first, otherwise AI will produce misleading results. Instead of being the solution, AI adds to the drug discovery problems.
Understanding Biomedical data
There are millions of scientific publications and thousands of patents about cancer. The data about cancer, being unstructured, is complex. Another challenge is the entanglement of the data. For instance, blocking the action of the vascular endothelial growth factor (VEGF), a signalling protein that promotes the growth of new blood vessels, is used for the treatment of several different cancers including colorectal cancer, glioblastoma, non-small cell lung cancer, ovarian cancer, fallopian tube cancer, peritoneal cancer, renal cell carcinoma and cervical cancer. At the same time each cancer can be treated with drugs acting on other targets. For example, colorectal cancer can be treated with agents targeting VEGFR2, PDGFRβ, RAF, KIT, RET and PD-1.
Biological Pathways
Another aspect of the entanglement is the biological pathways. Biological pathways consist of various bio-molecular interactions, e.g. protein-protein interactions, protein-DNA interactions, protein-RNA, etc. In cancer, the pathways do not work properly. Recent studies have found that different pathways can lead to the same cancer in different patients.
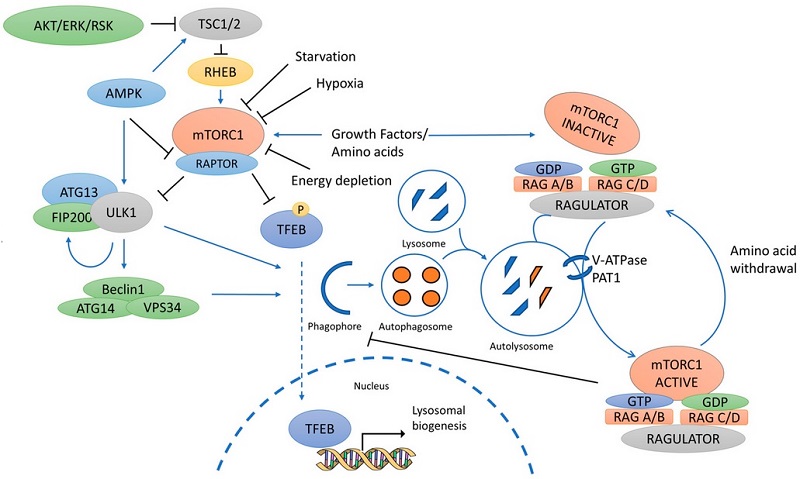
Data scientists and software engineers have limited understanding of how a biological pathway looks like. For this reason, I included Fig. 2 which shows mTOR pathway in cancer and autophagy. “mTOR” is a protein that serves as a central regulator of cell metabolism, growth, proliferation and survival. It interacts with at least 800 other proteins. Autophagy (or autophagocytosis) is the natural, regulated mechanism of the cell that disassembles unnecessary or dysfunctional components. There are hundreds pathway related to different aspects of cancer.
Fig. 2. mTOR pathway in cancer and autophagy
Another added complexity of biomedical data is that all types of cancer can share the same pathways responsible for tumor growth, cell migration, proliferation¸ autophagy, angiogenesis, etc. There is also interdependencies between the pathways. Migration and proliferation depend on growth, autophagy is influenced by the growth factors, etc.
Features Extraction in Small Molecules and Biologics
Manually extracting the relationship between different data sets and understanding the entanglement in the data is prohibitively difficult. It is beyond the ability of the human brain. According to human cognitive research, the human brain cannot process more than four variables at a time. Moreover, a researcher can only read a few of the millions of documents. With the right algorithms, computers have no limit. They provide reliable and repeatable results. Analytics and machine learning can provide powerful predictive models for all stages of cancer drug development.
Iteru Solution
There is no open source software for feature extraction that addresses the problems posed by biomedical data. The solution is to formulate proprietary algorithms. This what Iteru did. Iteru’s algorithms can be used on their own or in conjunction with open source feature extractors.
Iteru’s algorithms include the following:
- Tools to extract data based on the objective of analysis. The objective of analysis could be related to a combination of cancers or a specific cancer type, or could be narrowed to pathways, kinases, biomarkers, genes, proteins, etc.
- Selection of all biological entities and features related to the objective of analysis.
- Determine the hierarchical relationship between biological entities.
- Allow the user to interrogate the data to clarify ambiguity or verify the relevance of entities. Then the user can re-adjust feature extraction to ensure accuracy of AI results.

Как приобрести диплом техникума с минимальными рисками